The stunning beauty of Cell Biology - man's advance at the hand of Art
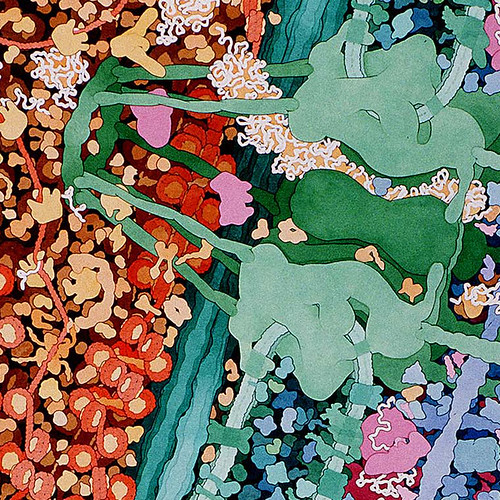
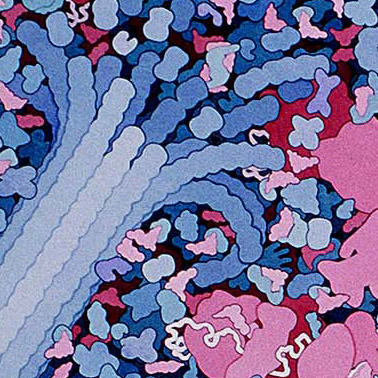
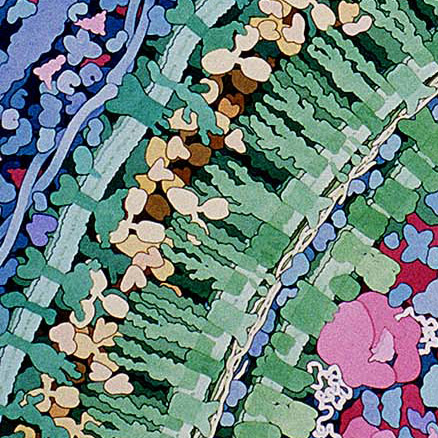
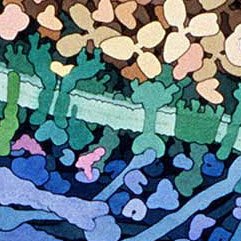
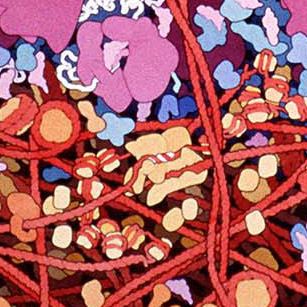
These amazing images are to-scale watercolor illustrations of a bacterium and blood cells. These are small sections of a larger piece made by Dr. David Goodsell. Notes on this work say
Macrophages circulate through the blood, searching for bacterial infection. When bacteria are found, macrophages engulf and digest them. This series of three paintings shows a macrophage engulfing a bacterium. Only a portion of the two cells, where a pseudopod of the macrophage is extending over the bacterium, is shown. The original paintings are 1 meter tall--at this magnification, the macrophage would fill most of a building.
David S. Goodsell is someone I admire deeply for his ability to illustrate Cell Biology (my favorite part of the life sciences, cant help the bias).
He is an associate professor in the Department of Molecular Biology at the Scripps Research Institute in La Jolla, California. He is the author of Bionanotechnology: Lessons from Nature (J. Wiley and Sons, 2004), Our Molecular Nature: The Body's Motors, Machines, and Messages (Springer-Verlag, 1996), and The Machinery of Life (Springer-Verlag, 1993). His recent biomolecular artwork can be viewed here. Address: Department of Molecular Biology, The Scripps Research Institute, La Jolla, CA 92037. Internet: goodsell@scripps.edu (I got this blurb from here)
Read about his scientific interests here.
Read about his artistic work here.
A nice online presentation he has on Overview of Biological Machines from the Designing Nanostructures: A tutorial site can be found here. And you can pull up a nice slide presentation here.
Here is a lovely poster on Molecular Machinery (PDF 5MB)








